The variations of:
- Essential vs non-essential genes
- Synthetic lethals vs suppressors
- Including the Lee, et al. transcription data
- Including the BIND protein-protein network
download
the Lee, et al. network without any genetic interactions here.
download
the BIND network without any genetic interactions here.
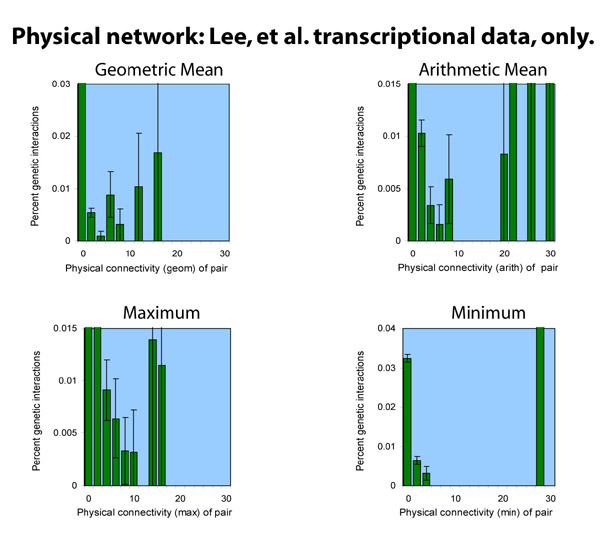
Including the Lee, et al. transcription data
In one recent version of the Lee, et al. transcription data, there
were 4,154 interactions with a p-value less than 10-3.
We combine that network with the MIPS and SGA genetic interaction
collections, producing the following analyses, analogous to figure
1(a) in our published paper:
download
the analysis in text form here.
download
the network here.
The transcription data is biased in its network topology; by definition, there are between one and two hundred hubs (the baits used in their data collection), and the rest of the nodes will be very likely to be leaves.
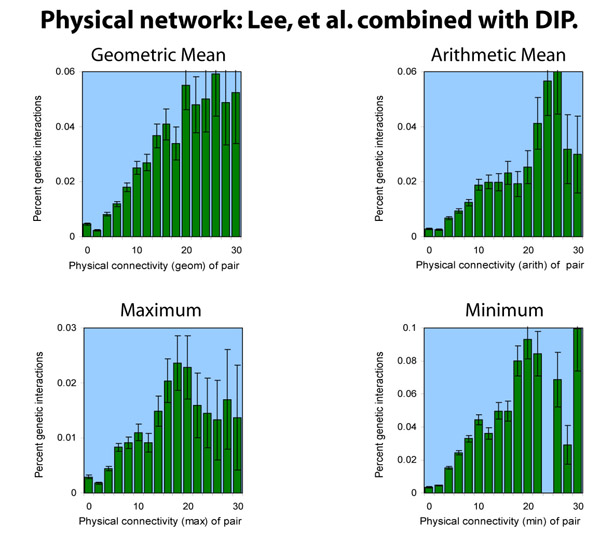
When the transcriptional network is combined with the DIP protein-protein interaction network, the results are quite similar to our original figure 1(a) analysis:
download
the analysis in text form here.
download
the network here.
This general result - trends that closely resemble the DIP-only trends - is not surprising given that the DIP network is much larger (15,114 interactions) than the Lee, et al. network (4,154 interactions).
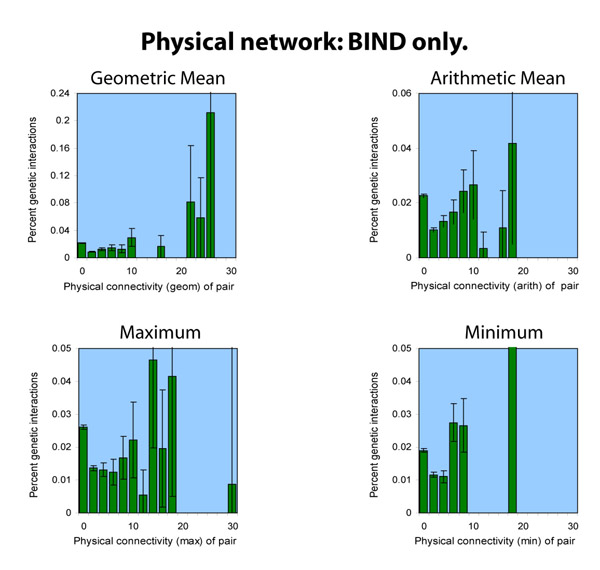
Including the BIND protein-protein data
The BIND database had 5,003 protein-protein interactions in it as
of our download; of those, almost all appear among the 15,114 DIP
interactions; only 210 do not. Thus, we don't consider BIND and DIP
together, since the result would be even less interesting than DIP
and Lee together. Below, we consider BIND against the MIPS and SGA
genetic interaction data sets:
download
the analysis in text form here.
download
the network here.
BIND and the Lee data set both have the problem that they are sparse networks, even compared to the somewhat sparse DIP dataset. Thus, the analyses appear even less persuasive towards any conclusion.